Overview
- Enzyme Kinetics theory
- for two molecules to react
- they must be within bond-forming distance
- possess enough kinetic energy to overcome the activation energy (Ea)
- factors that affect these two conditions will either decrease or increase the reaction rate
- temperature: causes an increase in kinetic energy
- concentration of reactants: increases probability of collisions
- for two molecules to react
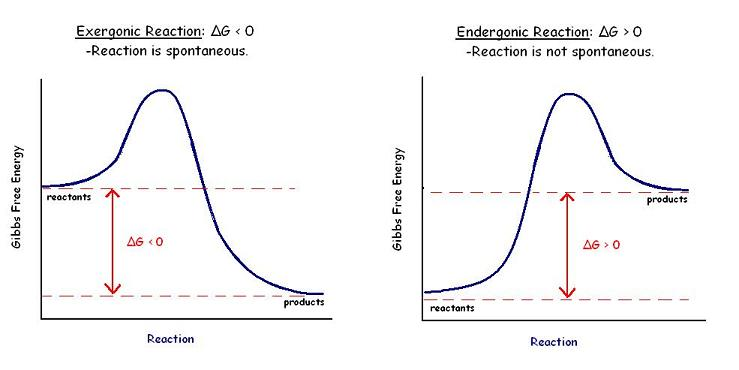
- Gibbs free energy change (ΔG)
- is the free energy change between the products and the reactants
- reflects the direction of a reaction and amount of reactants and products at equilibrium, but does NOT determine the rates of reaction
- ΔG < 0: reaction is spontaneous and favors product formation
- ΔG = 0: reaction is at equilibrium and proceeds in both direction at equal rates
- ΔG > 0: reaction is nonspontaneous and favors reactant formation
- Ea determines the rate of the reaction
- a large Ea will have a slower rate
- a small Ea will have a faster rate
- enzymes lower the Ea allowing the reaction to proceed at a faster rate
- enzymes do NOT change the ΔG of the reaction just the Ea
- enzymes are sensitive to temperature and pH
Enzymes Kinetics
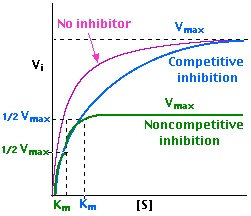
- Michaelis-Menten Equation
- an equation that relates the initial reaction velocity (Vi) to the substrate concentration
- Vmax is directly proportional to the [E]
- Km is the Michaelis-Menten constant which represents the substrate concentration at which Vi is half the maximum velocity (Vmax)
- Km = [S] at 1/2 Vmax
- Km is related to the enzyme’s affinity for the substrate [S]
- ↑ Km = ↓ affinity
- ↓ Km = ↑ affinity
- inhibitors affect these enzyme parameters
- an equation that relates the initial reaction velocity (Vi) to the substrate concentration
- Lineweaver-Burk Equation
- an inverted form of the Michaelis-Menten equation
- used to calculate Vmax and Km from experimental data at below enzyme saturation levels
- the equation is in the format y = ax + b (a is the slope and b is the y intercept)
- y = 1/Vi
- x = 1/[S]
- a = Km/Vmax
- b = 1/Vmax
- helpful hints
- the smaller value of -1/Km, the greater the Km
- an inverted form of the Michaelis-Menten equation
- ↑ y-intercept = ↓ Vmax
Enzyme kinetics is the study of the rates of enzymatic reactions and the factors that influence those rates. Enzymes are biological catalysts that accelerate chemical reactions in living organisms by lowering the activation energy required for the reactions to occur.
Enzyme kinetics provides a quantitative understanding of how enzymes work and how they interact with substrates and inhibitors. The kinetics of an enzyme-catalyzed reaction can be described by several parameters, including the rate of reaction, the Michaelis-Menten constant (Km), and the maximum reaction rate (Vmax).
The Michaelis-Menten equation is commonly used to describe the kinetics of enzyme-catalyzed reactions:
v = (Vmax [S]) / (Km + [S])
where:
- v is the initial rate of the reaction,
- [S] is the substrate concentration,
- Vmax is the maximum reaction rate when the enzyme is fully saturated with substrate,
- Km is the Michaelis-Menten constant, which represents the substrate concentration at which the reaction rate is half of Vmax.
The Michaelis-Menten equation assumes that the enzyme-substrate complex forms reversibly and that the rate of product formation is much slower than the rate of substrate binding and product release. It also assumes that the concentration of substrate is much higher than the concentration of enzyme.
Enzyme kinetics experiments often involve measuring the initial rate of the reaction at various substrate concentrations. By plotting the data and analyzing the resulting curve, parameters such as Km and Vmax can be determined.
Other important concepts in enzyme kinetics include enzyme inhibition, which can be competitive, non-competitive, or uncompetitive, and enzyme regulation, which involves the control of enzyme activity by factors such as allosteric regulation, covalent modification, or gene expression.
Enzyme kinetics is a fundamental field in biochemistry and provides insights into the mechanisms of enzymatic reactions, which have numerous applications in fields such as medicine, biotechnology, and drug development.
Enzyme Structure
Enzymes are proteins that serve as catalysts for biological reactions. They are made up of long chains of amino acids that fold into specific three-dimensional structures. The structure of an enzyme is critical to its function, as it determines the active site where the substrate binds and the catalytic reaction takes place.
The primary structure of an enzyme refers to the linear sequence of amino acids that make up the protein chain. The sequence is determined by the genetic information encoded in the DNA. The specific sequence of amino acids determines the overall shape and properties of the enzyme.
The secondary structure of an enzyme refers to the local folding patterns within the protein chain. The most common secondary structures in enzymes are alpha helices and beta sheets. These structures are stabilized by hydrogen bonding between the amino acid residues.
The tertiary structure of an enzyme is the overall three-dimensional arrangement of the protein chain. It is determined by various interactions, including hydrogen bonding, electrostatic interactions, hydrophobic interactions, and disulfide bonds. The tertiary structure brings together different secondary structural elements to form the functional enzyme.
The active site of an enzyme is a region within the enzyme’s three-dimensional structure where the substrate binds and the catalytic reaction occurs. The active site is often a small, well-defined pocket or cleft within the enzyme. It has a specific shape and chemical properties that complement the substrate, allowing for precise binding and catalysis.
In some cases, enzymes can have quaternary structure, which refers to the arrangement of multiple protein subunits to form a functional enzyme complex. Each subunit contributes to the overall structure and function of the enzyme complex.
Enzyme structure can be determined using techniques such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM). These methods provide detailed information about the arrangement of atoms within the enzyme and help in understanding its catalytic mechanism.
Overall, the structure of an enzyme is crucial for its function, as it determines the specificity and efficiency of catalysis. Changes in the enzyme structure, such as mutations or denaturation, can have significant effects on enzyme activity and can lead to various diseases and disorders.
Enzyme Kinetics Studies
- Determination of Reaction Rates: Enzyme kinetics experiments often involve measuring the rate of reaction at different substrate concentrations. By monitoring the formation of product over time, the initial rate of the reaction can be determined. This data is used to understand how the rate of the reaction changes with varying substrate concentrations.
- Michaelis-Menten Kinetics: The Michaelis-Menten equation is a fundamental tool in enzyme kinetics. It describes the relationship between the rate of reaction, substrate concentration, and key parameters such as Km and Vmax. Enzyme kinetics studies involve experimental determination of these parameters to understand the enzyme’s catalytic efficiency and affinity for the substrate.
- Lineweaver-Burk Plot: The Lineweaver-Burk plot is a graphical representation of the Michaelis-Menten equation. It allows for the determination of Km and Vmax by plotting the reciprocal of the reaction rate against the reciprocal of the substrate concentration. The slope and intercept of the line can be used to calculate these kinetic parameters.
- Enzyme Inhibition Studies: Enzyme kinetics can also involve studying the effects of inhibitors on the enzymatic reaction. Inhibition can be competitive, where the inhibitor competes with the substrate for the active site, or non-competitive, where the inhibitor binds to a different site on the enzyme. These studies provide insights into the regulation of enzyme activity and can be important in drug discovery and development.
- pH and Temperature Dependence: Enzyme kinetics studies often examine the effects of pH and temperature on enzyme activity. The pH can affect the ionization state of amino acid residues in the active site, thereby influencing substrate binding and catalysis. Temperature can affect the rate of the reaction by altering the enzyme’s conformation and the kinetic energy of the reactant molecules.
- Determination of Enzyme Kinetic Mechanisms: Enzyme kinetics studies can help elucidate the underlying kinetic mechanism of an enzyme-catalyzed reaction. This involves investigating how different substrates, products, and inhibitors influence the reaction rate. By studying the patterns of inhibition and substrate/product binding, researchers can infer the order of substrate binding and product release, as well as the presence of multiple substrate or product binding sites.
Check out Ultimate USMLE Step 1 Study Notes.